
How To Create a Mask File
Source:vignettes/tutorial-create-mask-file.Rmd
tutorial-create-mask-file.RmdThe Sparse Marginal Epistasis test (SME) requires a HDF5 file containing binary masking data. The effectiveness of the test relies on how informative the mask is for a specific trait. For instance, to test for marginal epistasis related to transcriptional activity, DNase I hypersensitivity sites data can be used to condition the detection on accessible chromatin regions.
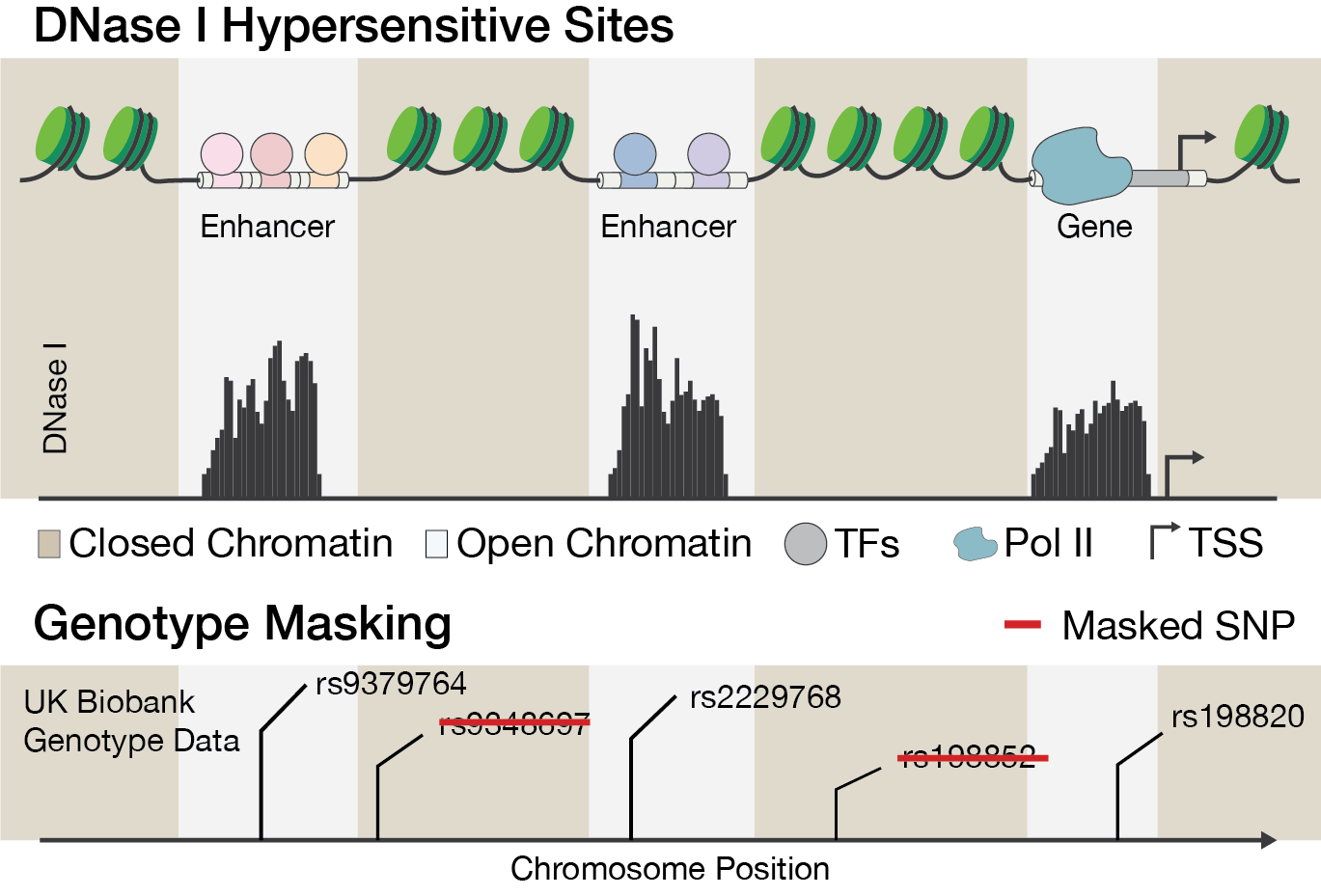 Figure 1. The illustration shows open chromatin data.
DNase I hypersensitive sites indicate transcriptional activity. The
DNase-seq signal is converted to a binary mask, where genetic variants
in inaccessible chromatin regions are excluded from the covariance
matrix for marginal epistasis.
Figure 1. The illustration shows open chromatin data.
DNase I hypersensitive sites indicate transcriptional activity. The
DNase-seq signal is converted to a binary mask, where genetic variants
in inaccessible chromatin regions are excluded from the covariance
matrix for marginal epistasis.
Mask File Format
The sme() function expects the mask data to be in an
HDF5 file. The HDF5 format includes two primary object types:
- Datasets - typed multidimensional arrays
- Groups - container structures that can hold datasets and other groups
Mask Format Requirements
The mask data should be organized into the following groups and datasets:
Groups:
-
ld: Contains SNPs in linkage disequilibrium (LD) with the focal SNP, which will be excluded. -
gxg: Contains indices of SNPs used to condition the marginal epistasis test, which will be included.
The required group names can be configured as input parameters of
sme(). The defaults are ld and
gxg.
Datasets:
-
ld/<j>: For each focal SNP<j>, this dataset contains indices of SNPs in the same LD block as that SNP. These SNPs will be excluded from the gene-by-gene interaction covariance matrix. -
gxg/<j>: For each focal SNP<j>, this dataset contains indices of SNPs to include in the the gene-by-gene interaction covariance matrix for focal SNP<j>.
Important: All indices in the mask file must be
zero-based and should correspond to the zero-based row
indices of the PLINK .bim file. This includes the dataset
index (<j> in gxg/<j>) and the
data itself. This zero-based indexing is necessary because the mask data
is read by a C++ subroutine in sme(), which uses zero-based
indexing, unlike R’s one-based indexing for SNP indices in the function
call.
Creating and Using Mask Files
The package provides utility functions to create, write, and read
valid mask files for sme().
hdf5_file <- tempfile()
# Group names
gxg_h5_group <- "gxg"
ld_h5_group <- "ld"
# Data (still in 1-based R indexing)
include_gxg_snps <- 1:10
exclude_ld_snps <- 5:6
# Focal SNP (still in 1-based R indexing)
focal_snp <- 4
# Dataset names
dataset_name_pattern <- "%s/%s"
# 0-based index!
gxg_dataset <- sprintf(dataset_name_pattern, gxg_h5_group, focal_snp - 1)
ld_dataset <- sprintf(dataset_name_pattern, ld_h5_group, focal_snp - 1)
# Create an empty HDF5 file
create_hdf5_file(hdf5_file)
# Write LD data
write_hdf5_dataset(hdf5_file, ld_dataset, exclude_ld_snps - 1) # 0-based index!
# Write GXG data
write_hdf5_dataset(hdf5_file, gxg_dataset, include_gxg_snps - 1)We can check that the data was written correctly.
ld_read <- read_hdf5_dataset(hdf5_file, ld_dataset)
gxg_read <- read_hdf5_dataset(hdf5_file, gxg_dataset)
print(sprintf("Zero-based indices of SNPs to exclude: %s", str(ld_read)))
#> int [1:2] 4 5
#> character(0)
print(sprintf("Zero-based indices of SNPs to include: %s", str(gxg_read)))
#> int [1:10] 0 1 2 3 4 5 6 7 8 9
#> character(0)SessionInfo
sessionInfo()
#> R version 4.5.1 (2025-06-13)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.2 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] smer_0.0.2
#>
#> loaded via a namespace (and not attached):
#> [1] jsonlite_2.0.0 dplyr_1.1.4 compiler_4.5.1
#> [4] tidyselect_1.2.1 Rcpp_1.1.0 FMStable_0.1-4
#> [7] parallel_4.5.1 tidyr_1.3.1 jquerylib_0.1.4
#> [10] systemfonts_1.2.3 textshaping_1.0.1 harmonicmeanp_3.0.1
#> [13] yaml_2.3.10 fastmap_1.2.0 R6_2.6.1
#> [16] generics_0.1.4 knitr_1.50 genio_1.1.2
#> [19] iterators_1.0.14 backports_1.5.0 checkmate_2.3.3
#> [22] tibble_3.3.0 desc_1.4.3 bslib_0.9.0
#> [25] pillar_1.11.0 rlang_1.1.6 cachem_1.1.0
#> [28] xfun_0.53 fs_1.6.6 sass_0.4.10
#> [31] cli_3.6.5 pkgdown_2.1.3 magrittr_2.0.3
#> [34] digest_0.6.37 mvMAPIT_2.0.3 foreach_1.5.2
#> [37] mvtnorm_1.3-3 lifecycle_1.0.4 CompQuadForm_1.4.4
#> [40] vctrs_0.6.5 evaluate_1.0.4 glue_1.8.0
#> [43] codetools_0.2-20 ragg_1.4.0 purrr_1.1.0
#> [46] rmarkdown_2.29 tools_4.5.1 pkgconfig_2.0.3
#> [49] htmltools_0.5.8.1